
Loghman Samani
Master’s Student in Technical Biology at The University of Stuttgart
Email: st186417@stud.uni-stuttgart.de
I have a strong academic foundation in Cell and Molecular Biology, with growing expertise in computational biology and machine learning.
About Me
I am a master’s student in Computational Biology at the University of Stuttgart, currently working on my master’s thesis titled: “Designing Gene Regulatory Networks through Evolutionary and Gradient-Based Optimization: A Computational Approach to Predicting Spatial Patterns in 2D Reaction-Diffusion Systems."
With a background in Cell and Molecular Biology (B.Sc. from the University of Kurdistan), I am passionate about applying computational methods to explore biological systems at the molecular level. My focus is on leveraging these methods to enhance our understanding and description of complex biological processes.
In addition to my core studies, I have a deep interest in artificial intelligence, particularly in using deep learning to automate and accelerate the analysis of such intricate systems. Alongside my thesis, I actively engage in mini-projects that further expand my skills in AI and machine learning.
Beyond computational biology, I am developing a growing interest in cognitive neuroscience, eager to explore how these fields intersect and contribute to our understanding of the brain and human cognition.
Achievements
Cell and Molecular Biology Olympiad
I ranked 11th in the final stage of the Cell and Molecular Biology Olympiad at Iran's national scientific competition.
Member of the Elite Foundation
In 2018, after achieving 14th place in the national Master's exam in Biophysics and 11th in the Cell and Molecular Biology Olympiad, I had the honor of being inducted as a member of the Elite Students Foundation at the University of Kurdistan.
Projects
Deep-Learning Models From Scratch
Feb 2024 - Sep 2024

Over eight months, I embarked on a comprehensive journey into deep learning by studying the
Deep Learning Specialization from
deeplearning.ai on Coursera. To fully grasp the fundamentals and intricacies of these models,
I implemented over 15 different neural networks, mostly from scratch. These implementations gave me a deep, hands-on understanding of how various models function and the
underlying principles of deep learning.
Implemented Models and Techniques:
- Multi-Layer Perceptrons (MLPs): Various architectures for different tasks, demonstrating a thorough grasp of fully connected layers.
- Convolutional Neural Networks (CNNs): Explored image recognition and processing with CNNs, enhancing feature extraction and accuracy.
- Recurrent Neural Networks (RNNs) & LSTMs: Worked on sequence prediction tasks using RNNs and Long Short-Term Memory networks, mastering time-series data handling.
- Transformers: Implemented transformers to understand attention mechanisms and apply them to NLP tasks.
- Regularization Methods: Experimented with L2 regularization, dropout, and other techniques to reduce overfitting and improve model generalization.
- Optimization Algorithms: Implemented Gradient Descent (GD), Stochastic Gradient Descent (SGD), Batch Gradient Descent, and Adam to explore different approaches to model training.
Explore the project on GitHub.
Biostoch: A Python Library for Simulating Biological Systems
2023 - Ongoing Development
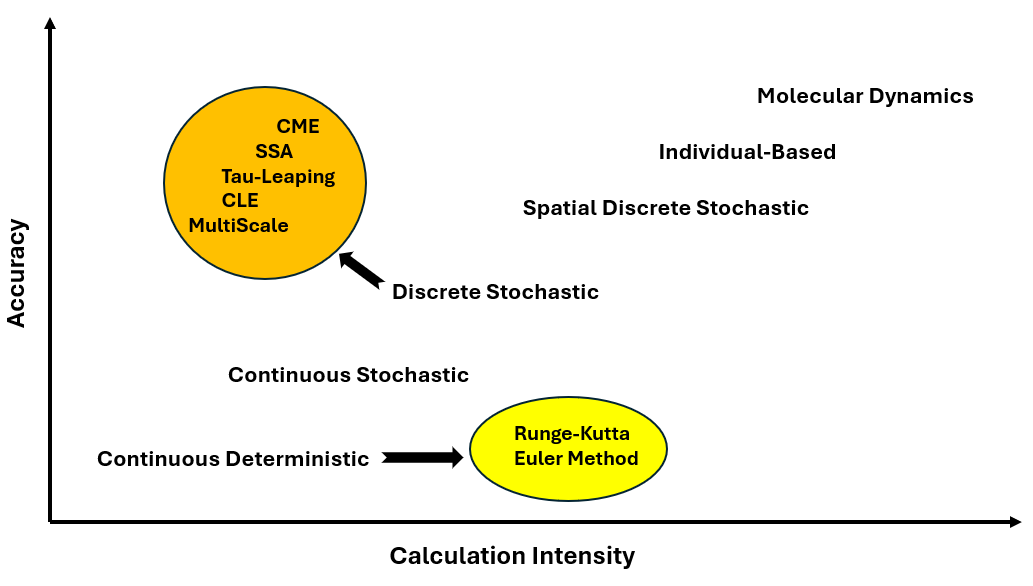
Biostoch is a versatile Python library designed for simulating biological systems using both deterministic and stochastic methods.
Whether you are modeling cellular processes or biochemical reactions, Biostoch provides a range of algorithms to capture the dynamics of these systems with high accuracy.
Currently, the library includes:
Deterministic Algorithms:
- Euler's Method: A simple numerical approach for solving ordinary differential equations by approximating values at discrete intervals.
- Runge-Kutta Method: A more advanced and accurate algorithm for solving differential equations, offering improved precision with intermediate steps.
Stochastic Algorithms:
- Stochastic Simulation Algorithm (SSA): Models random fluctuations in biochemical reactions, capturing inherent randomness.
- Tau-Leaping Algorithm: Accelerates simulations by approximating system changes over larger time steps.
- Chemical Langevin Equation (CLE): Combines both deterministic and stochastic elements to model complex chemical reactions.
Currently, Biostoch is optimized for one-dimensional simulations of biological systems, such as individual cell modeling. Future updates will expand its capabilities to support two-dimensional and three-dimensional simulations.
Explore the project on GitHub.
SmartSolve
Jan 2023 - Jan 2024

SmartSolve is a Python library designed to streamline the use of classical machine learning models. It is structured into three key components: preprocessing, models, and evaluation. The library encompasses 12 diverse machine learning algorithms, including:
- Linear Regression
- Logistic Regression
- Decision Trees
- Random Forests
- K-Means Clustering
- K-Nearest Neighbors
- Principal Component Analysis
- Support Vector Machines
- Gradient Boosting
- Singular Value Decomposition
- Gaussian Mixture Models
- Naive Bayes
Additionally, SmartSolve provides tools for data preprocessing and model performance evaluation. For more information, read the article Introducing SmartSolve: A Mini Machine Learning Package on Medium. This article offers an overview of the library’s features, including preprocessing tools, model implementations, and evaluation methods.
Explore the project on GitHub.
MAPK Signaling Pathway
Jun 2023 - Dec 2023
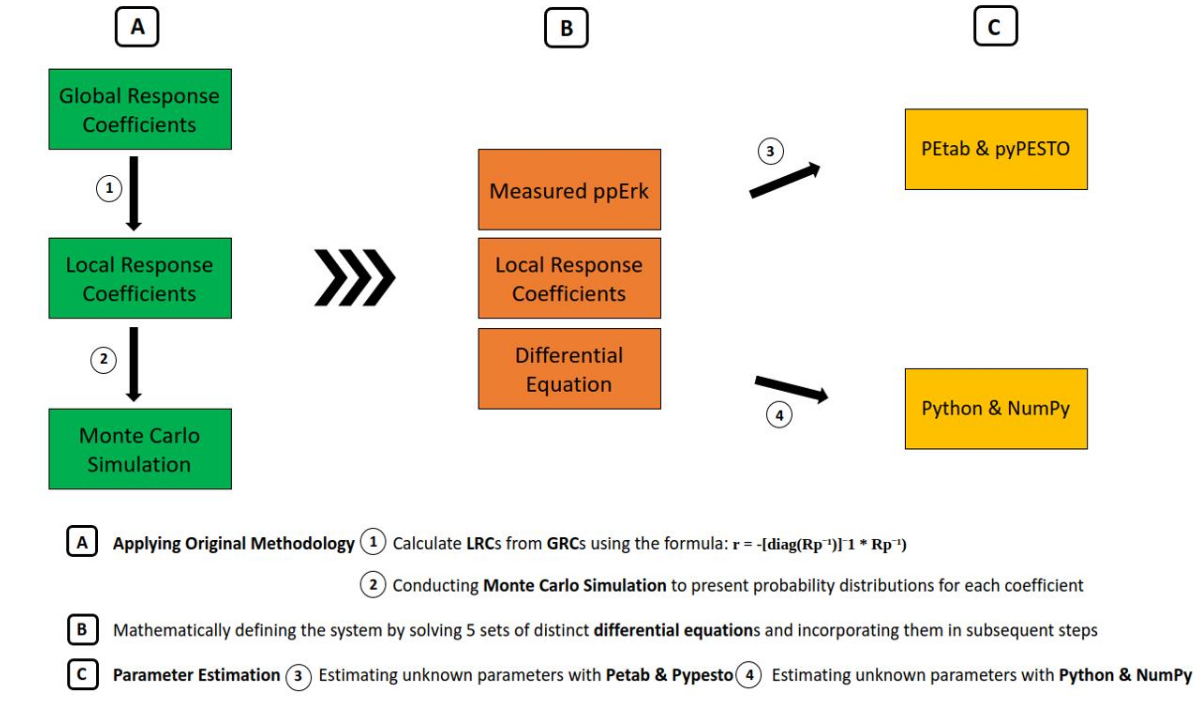
This project focuses on replicating the study presented in the article titled Growth Factor-Induced MAPK Network Topology Shapes Erk Response Determining PC-12 Cell Fate by Silvia D. M. Santos, Peter J. Verveer, and Philippe I. H. Bastiaens, published in 2007. Our objective was to reproduce the findings and methodologies outlined in this influential paper.
In addition to replication, we have redefined various aspects of the MAPK signaling pathway and utilized different tools and parameter estimation methods to uncover previously unknown parameters.
For a detailed account of our work, you can read the paper Unraveling the Complexity of MAPK Signaling Pathway.
Explore the project on GitHub.