
Loghman Samani
I hold a master degree in Computational Biology from the University of Stuttgart.
My background bridges molecular biology and deep learning, with a focus on generative models and their applications in structural biology.
About Me
I recently completed my master’s thesis at the University of Stuttgart, titled “Automated Design and Analysis of Gene Regulatory Networks (GRNs)”. Through this work, I developed a hybrid optimization framework for designing Gene Regulatory Networks, combining evolutionary algorithms with gradient-based methods to simulate realistic biological systems. Currently, I am seeking a Ph.D. position in computational biology, with a focus on structural biology. My academic journey began with a B.Sc. in Cell and Molecular Biology at the University of Kurdistan, where I cultivated a strong foundation in biological sciences. Over time, I expanded my expertise by integrating computational tools and machine learning techniques to investigate molecular systems and unravel complex biological processes. Beyond my primary research interests, I have a keen fascination with artificial intelligence, particularly its applications in simplifying and accelerating the analysis of intricate biological systems. To further enhance my skills, I actively engage in side projects related to AI and machine learning. Recently, I have also delved into cognitive neuroscience, exploring its intersection with computational biology and gaining a deeper appreciation for understanding the brain and human cognition from a multidisciplinary perspective.
Achievements
Cell and Molecular Biology Olympiad
I ranked 11th in the final stage of the Cell and Molecular Biology Olympiad at Iran's national scientific competition.
Member of the Elite Foundation
In 2018, after achieving 14th place in the national Master's exam in Biophysics and 11th in the Cell and Molecular Biology Olympiad, I had the honor of being inducted as a member of the Elite Students Foundation at the University of Kurdistan.
Projects
A high-performance C++ deep learning library for building and training neural networks
June 2025 - Present

CppNet is a high-performance C++ deep learning library for building and training neural networks from scratch.
Designed for speed and flexibility, CppNet combines Eigen for efficient tensor operations, OpenMP for CPU parallelism, and CUDA for GPU acceleration.
The library features a clean modular API with clearly separated components for layers, losses, optimizers, metrics, and regularizations, making it easy to extend with custom modules.
The project follows a clear structure with include/ headers (public API), src/ implementations, and examples/ for practical demonstrations.
Explore the project on GitHub.
A PyTorch-based library for diffusion models
May 2025 - Present

In TorchDiff, I developed a comprehensive PyTorch-based library for building, experimenting with, and deploying diffusion models, drawing inspiration from recent advancements in generative AI. The library currently implements five major diffusion model families:
- Denoising Diffusion Probabilistic Models (DDPM)
- Denoising Diffusion Implicit Models (DDIM)
- Score-based / SDE-driven diffusion processes
- Latent Diffusion Models (LDM)
- UnCLIP, the hierarchical text-conditional image-generation framework
For more details, visit the project homepage
See the full implementation on GitHub.
Automated Design and Analysis of Gene Regulatory Networks (GRNs)
March 2024 - February 2025
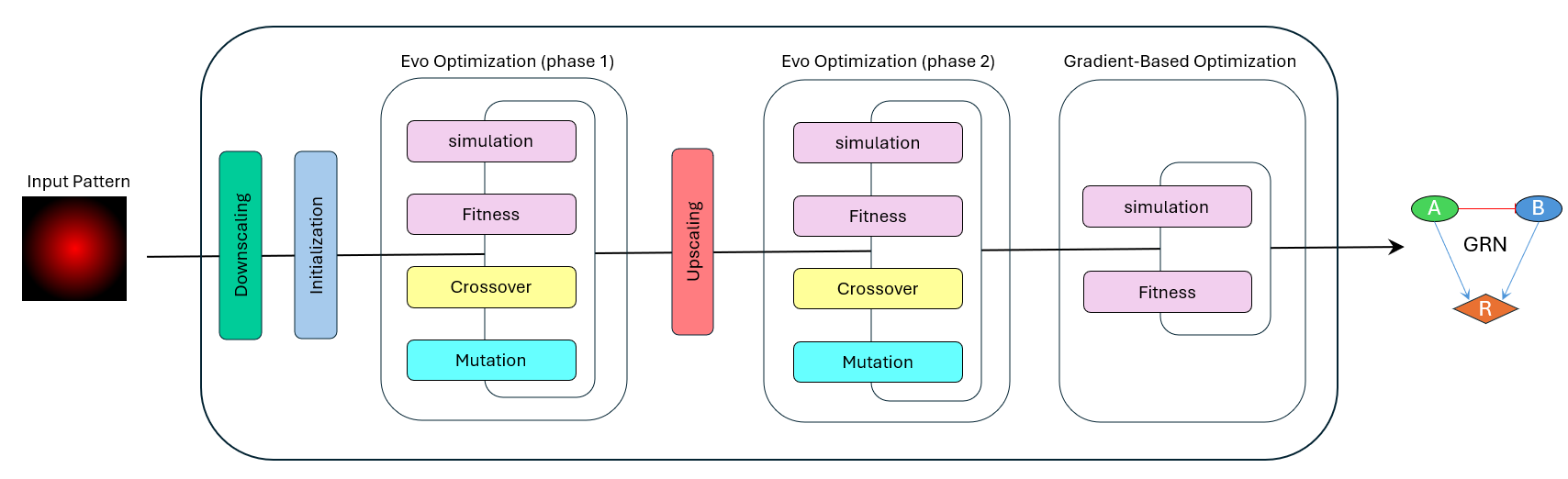
As part of my master’s thesis at the University of Stuttgart—Institute for Stochastics and Applications, I developed the GRN-Designer algorithm, a novel computational framework for designing and analyzing Gene Regulatory Networks (GRNs). GRNs serve as blueprints for understanding how genes interact and regulate each other in biological systems. While traditional GRN models often assume uniform spatial distributions of genes, this assumption does not always reflect real-world scenarios. The GRN-Designer algorithm addresses this limitation by employing a hybrid optimization approach that combines evolutionary algorithms (inspired by natural selection) with gradient-based methods (for fine-tuning). This enables the algorithm to design GRNs capable of recreating predefined spatial patterns. A key innovation of this framework is its ability to simultaneously optimize both the initial gene expression levels and the spatial activation potential of genes, simulating how gene activity propagates across a two-dimensional space. To validate the designs, I utilized partial differential equations (PDEs) to model the dynamics of gene expression accurately. The algorithm successfully generated GRNs for a range of spatial patterns, from simple gradients to complex shapes, while dynamically adjusting the complexity of the network to match the intricacy of the target pattern. This work has significant implications for synthetic biology and tissue engineering, where precise spatial patterning is crucial. Future developments could extend the framework to three-dimensional environments, incorporate stochastic elements to better mimic biological variability, and address even more sophisticated systems.
For more details, visit the project homepage.
Deep-Learning Models From Scratch
Feb 2024 - Sep 2024

Over eight months, I completed the Deep Learning Specialization by deeplearning.ai on Coursera, implementing over 15 neural networks from scratch. This hands-on approach deepened my understanding of fundamental models such as Multi-Layer Perceptrons (MLPs), Convolutional Neural Networks (CNNs), Recurrent Neural Networks (RNNs) with LSTMs, and Transformers. I also explored regularization techniques like L2 regularization and dropout, as well as optimization algorithms including Gradient Descent, SGD, and Adam. By building these models from the ground up, I gained practical insights into their inner workings and applications in tasks like image recognition, sequence prediction, and natural language processing. This project reflects my commitment to mastering deep learning through rigorous study and implementation.
Explore the project on GitHub.
Biostoch: A Python Library for Simulating Biological Systems
2023 - Ongoing Development
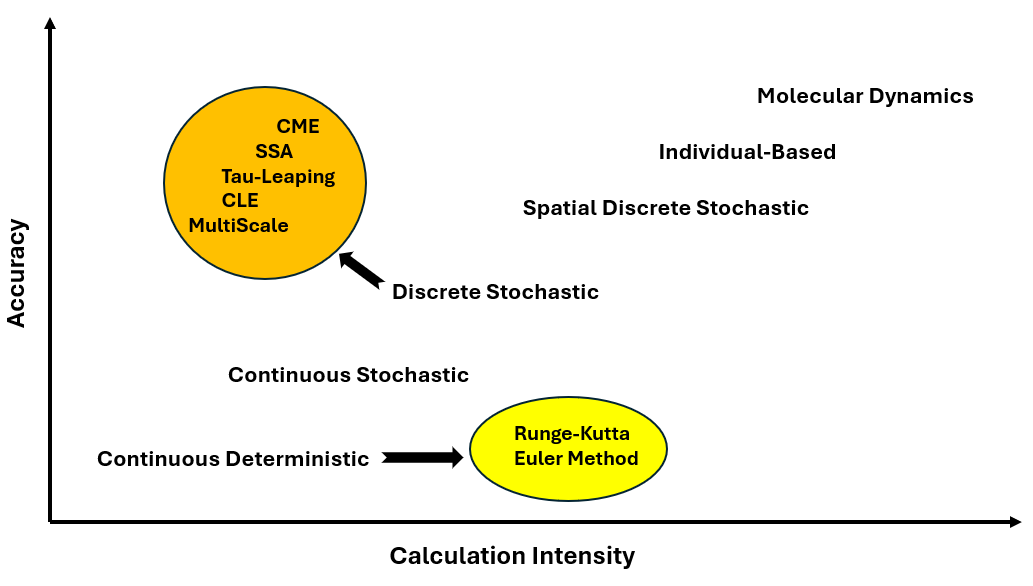
Biostoch is a Python library designed for simulating biological systems using both deterministic and stochastic methods, enabling researchers to model cellular processes and biochemical reactions with high accuracy. The library currently supports one-dimensional simulations and includes a range of algorithms: deterministic methods such as Euler's Method and the Runge-Kutta Method for solving ordinary differential equations, and stochastic methods like the Stochastic Simulation Algorithm (SSA), Tau-Leaping Algorithm, and Chemical Langevin Equation (CLE) for capturing random fluctuations in biochemical reactions. By combining these approaches, Biostoch provides flexibility in modeling systems with varying levels of complexity and computational demands. Future updates will expand its capabilities to support two-dimensional and three-dimensional simulations, further enhancing its utility for studying multi-scale biological phenomena.
Explore the project on GitHub.
SmartSolve
Jan 2023 - Jan 2024

SmartSolve is a Python library designed to simplify the implementation and evaluation of classical machine learning models. It provides a streamlined workflow for data preprocessing, model training, and performance assessment, making it an ideal tool for both beginners and experienced practitioners in the field of machine learning.
The library is organized into three core components: preprocessing, models, and evaluation. These modules work seamlessly together to facilitate efficient experimentation with various algorithms. SmartSolve supports a wide range of machine learning techniques, including:
- Regression Models: Linear Regression, Logistic Regression
- Tree-Based Models: Decision Trees, Random Forests
- Clustering Algorithms: K-Means Clustering, Gaussian Mixture Models
- Dimensionality Reduction Techniques: Principal Component Analysis, Singular Value Decomposition
- Classification Algorithms: Support Vector Machines, Naive Bayes, K-Nearest Neighbors
- Ensemble Methods: Gradient Boosting
Beyond its extensive collection of algorithms, SmartSolve includes robust tools for data preprocessing (e.g., feature scaling, encoding categorical variables) and model evaluation (e.g., cross-validation, confusion matrices, ROC curves). These features empower users to focus on solving real-world problems without getting bogged down in tedious setup tasks.
For a deeper dive into SmartSolve's capabilities, check out my article Introducing SmartSolve: A Mini Machine Learning Package on Medium. This article provides an overview of the library's design philosophy, key features, and practical use cases.
Explore the project on GitHub.
MAPK Signaling Pathway
Jun 2023 - Dec 2023
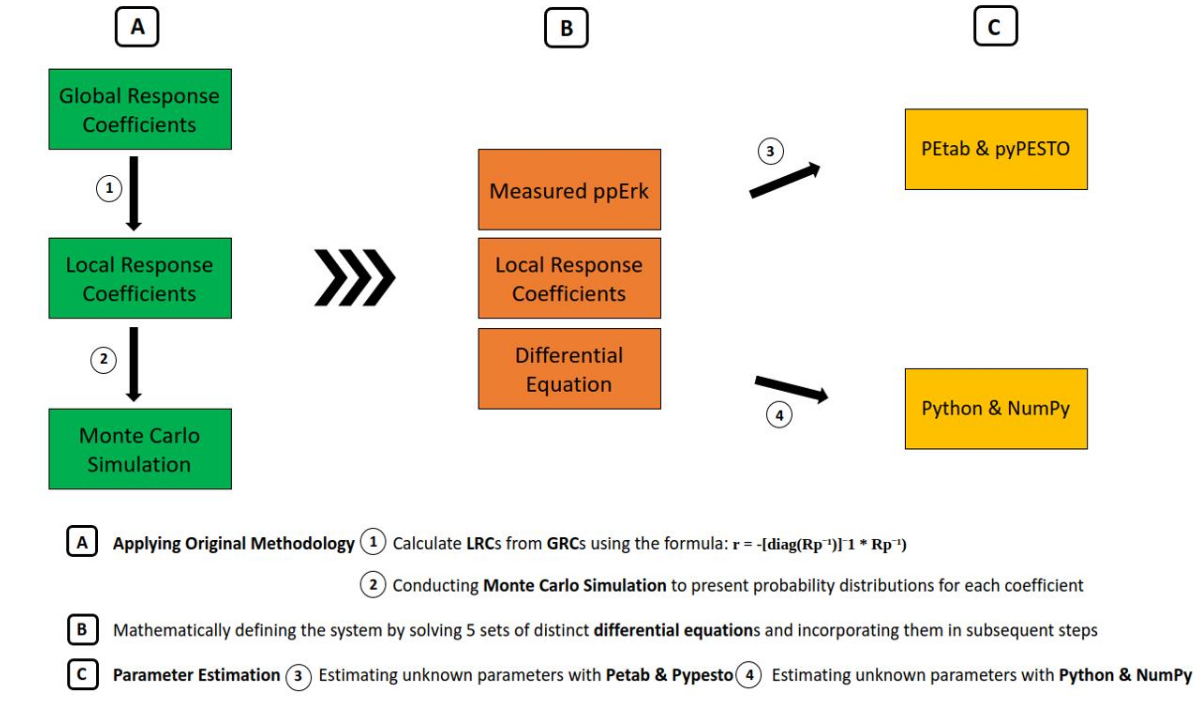
This project aimed to replicate and extend the study presented in the article Growth Factor-Induced MAPK Network Topology Shapes Erk Response Determining PC-12 Cell Fate, authored by Silvia D. M. Santos, Peter J. Verveer, and Philippe I. H. Bastiaens (2007). The MAPK signaling pathway plays a critical role in regulating cellular processes such as proliferation, differentiation, and apoptosis. Our work focused on reproducing the original findings while exploring new dimensions of this complex biological system.
In addition to replicating the methodologies outlined in the original study, we redefined key aspects of the MAPK signaling pathway using advanced computational tools and parameter estimation techniques. By doing so, we uncovered previously unknown parameters and gained deeper insights into the dynamics of the Erk response and its influence on PC-12 cell fate. These efforts contribute to a better understanding of how network topology shapes signaling outcomes in biological systems.
For a detailed exploration of our findings, you can read the paper Unraveling the Complexity of MAPK Signaling Pathway. This document provides an in-depth analysis of our approach, results, and implications for future research.
Explore the project on GitHub.